README.md
In mixOmicsTeam/mixOmics: Omics Data Integration Project

This repository contains the R package which is hosted on
Bioconductor
and our stable and development GitHub versions.
Installation
(macOS users only: Ensure you have installed
XQuartz first.)
From Bioconductor
The best way to install mixOmics is using Bioconductor. You can see
the landing page for the release version of mixOmics on Bioconductor
here.
Make sure you have the latest R version and the latest BiocManager
package installed following these
instructions.
## install BiocManager if not installed
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
## install mixOmics
BiocManager::install('mixOmics')
## load mixOmics
library(mixOmics)
From Github
Bioconductor versions are updated twice a year, between these updates
you can downlod the latest stable version of mixOmics from Github
using:
BiocManager::install('mixOmicsTeam/mixOmics')
You can also install the development
version for
new features yet to be widely tested:
BiocManager::install("mixOmicsTeam/mixOmics@development")
From Docker container
You can install our latest stable Github version of mixOmics via our
Docker container. You can do this by downloading and using the Docker
desktop application or via the command line as described below.
Click to expand
Note: this requires root privileges
1) Install Docker following instructions at
https://docs.docker.com/docker-for-mac/install/
if your OS is not compatible with the latest version download an
older version of Docker from the following link:
- MacOS: https://docs.docker.com/docker-for-mac/release-notes/
- Windows: https://docs.docker.com/docker-for-windows/release-notes/
Then open your system’s command line interface (e.g. Terminal for MacOS
and Command Promot for Windows) for the following steps.
MacOS users only: you will need to launch Docker Desktop to activate
your root privileges before running any docker commands from the command
line.
2) Pull mixOmics container
docker pull mixomicsteam/mixomics
3) Ensure it is installed
The following command lists the running images:
docker images
This lists the installed images. The output should be something similar
to the following:
$ docker images
> REPOSITORY TAG IMAGE ID CREATED SIZE
> mixomicsteam/mixomics latest e755393ac247 2 weeks ago 4.38GB
4) Activate the container
Running the following command activates the container. You must change
your_password to a custom password of your own. You can also customise
ports (8787:8787) if desired/necessary. see
https://docs.docker.com/config/containers/container-networking/ for
details.
docker run -e PASSWORD=your_password --rm -p 8787:8787 mixomicsteam/mixomics
5) Run
In your web browser, go to http://localhost:8787/ (change port if
necessary) and login with the following credentials:
username: rstudio
password: (your_password set in step 4)
6) Inspect/stop
The following command lists the running containers:
sudo docker ps
The output should be something similar to the following:
$ sudo docker ps
> CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
> f14b0bc28326 mixomicsteam/mixomics "/init" 7 minutes ago Up 7 minutes 0.0.0.0:8787->8787/tcp compassionate_mestorf
The listed image ID can then be used to stop the container (here
f14b0bc28326)
docker stop f14b0bc28326
Contribution
We welcome community contributions concordant with our code of
conduct.
We strongly recommend adhering to Bioconductor’s coding
guide for
software consistency if you wish to contribute to mixOmics R codes.
Bug reports and pull requests
To report a bug (or offer a solution for a bug!) visit:
https://github.com/mixOmicsTeam/mixOmics/issues. We fully welcome and
appreciate well-formatted and detailed pull requests. Preferably with
tests on our datasets.
Set up development environment
- Install the latest version of R
- Install RStudio
- Clone this repo, checkout master branch, pull origin and then run:
install.packages("renv", Ncpus=4)
install.packages("devtools", Ncpus=4)
# restore the renv environment
renv::restore()
# or to initialise renv
# renv::init(bioconductor = TRUE)
# update the renv environment if needed
# renv::snapshot()
# test installation
devtools::install()
devtools::test()
# complete package check (takes a while)
devtools::check()
Discussion forum
We wish to make our discussions transparent so please direct your
analysis questions to our discussion forum
https://mixomics-users.discourse.group. This forum is aimed to host
discussions on choices of multivariate analyses, as well as comments and
suggestions to improve the package. We hope to create an active
community of users, data analysts, developers and R programmers alike!
Thank you!
About the mixOmics team
mixOmics is collaborative project between Australia (Melbourne),
France (Toulouse), and Canada (Vancouver). The core team includes
Kim-Anh Lê Cao - https://lecao-lab.science.unimelb.edu.au (University
of Melbourne), Florian Rohart - http://florian.rohart.free.fr
(Toulouse) and Sébastien Déjean -
https://perso.math.univ-toulouse.fr/dejean/. We also have key
contributors, past (Benoît Gautier, François Bartolo) and present (Al
Abadi, University of Melbourne) and several collaborators including
Amrit Singh (University of British Columbia), Olivier Chapleur (IRSTEA,
Paris), Antoine Bodein (Universite de Laval) - it could be you too, if
you wish to be involved!.
The project started at the Institut de Mathématiques de Toulouse in
France, and has been fully implemented in Australia, at the University
of Queensland, Brisbane (2009 – 2016) and at the University of
Melbourne, Australia (from 2017). We focus on the development of
computational and statistical methods for biological data integration
and their implementation in mixOmics.
Why this toolkit?
mixOmics offers a wide range of novel multivariate methods for the
exploration and integration of biological datasets with a particular
focus on variable selection. Single ’omics analysis does not provide
enough information to give a deep understanding of a biological system,
but we can obtain a more holistic view of a system by combining multiple
’omics analyses. Our mixOmics R package proposes a whole range of
multivariate methods that we developed and validated on many biological
studies to gain more insight into ’omics biological studies.
Want to know more?
www.mixOmics.org (tutorials and resources)
Our latest bookdown vignette:
https://mixomicsteam.github.io/mixOmics-Vignette/
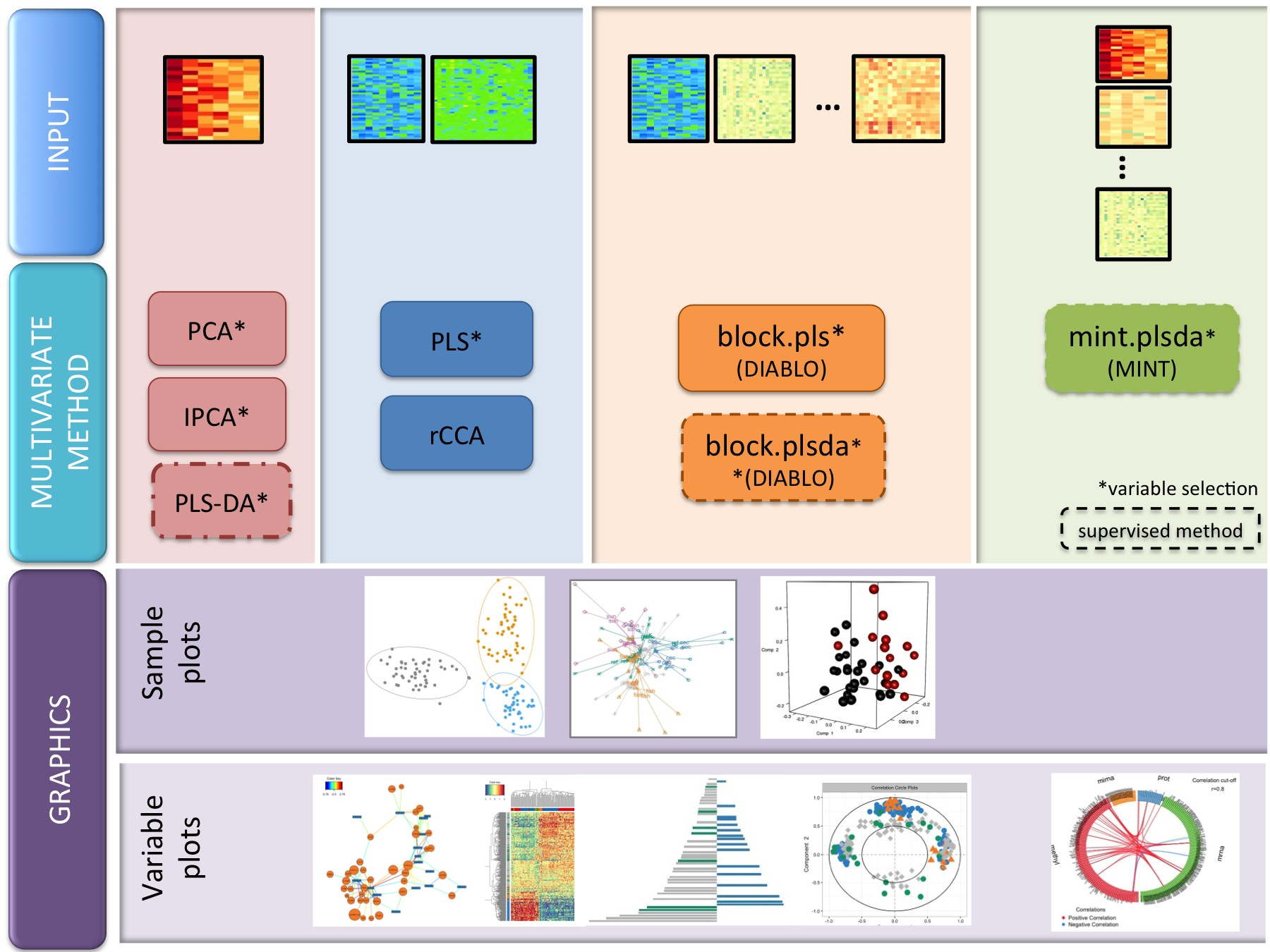
Different types of methods
We have developed 17 novel multivariate methods (the package includes 19
methods in total). The names are full of acronyms, but are represented
in this diagram. PLS stands for Projection to Latent Structures
(also called Partial Least Squares, but not our preferred nomenclature),
CCA for Canonical Correlation Analysis.
That’s it! Ready! Set! Go!
Thank you for using mixOmics!
What’s New
March 2022
- bug fix implemented for Issue
#196.
perf()
can now handle features with a (s)pls which have near zero variance.
- bug fix implemented for Issue
#192.
predict() can now handle when the testing and training data have
their columns in different orders.
- bug fix implemented for Issue
#178. If the
indY parameter is used in block.spls(), circosPlot() can now
properly identify the $Y$ dataframe.
- bug fix implemented for Issue
#172.
perf()
now returns values for the choice.ncomp component when nrepeat
$< 3$ whereas before it would just return NAs.
- bug fix implemented for Issue
#171.
cim()
now can take pca objects as input.
- bug fix implemented for Issue
#161.
tune.spca() can now handle NA values appropriately.
- bug fix implemented for Issue
#150. Provided
users with a specific error message for when
plotArrow() is run on a
(mint).(s)plsda object.
- bug fix implemented for Issue
#122. Provided
users with a specific error message for when a
splsda object that
has only one sample associated with a given class is passed to
perf().
- bug fix implemented for Issue
#120.
plotLoadings() now returns the loading values for features from
all dataframes rather than just the last one when operating on a
(mint).(block).(s)plsda object.
- bug fix implemented for Issue
#43. Homogenised
the way in which
tune.mint.splsda() and perf.mint.splsda()
calculate balanced error rate (BER) as there was disparity between
them. Also made the global BER a weighted average of BERs across each
study.
- enhancement implemented for Issue
#30/#34. The
parameter
verbose.call was added to most of the methods. This
parameter allows users to access the specific values input into the
call of a function from its output.
- bug fix implemented for Issue
#24.
background.predict() can now operate on mint.splsda objects and
can be used as part of plotIndiv().
July 2021
- new function
plotMarkers to visualise the selected features in block
analyses (see https://github.com/mixOmicsTeam/mixOmics/issues/134)
tune.spls now able to tune the selected variables on both X and
Y. See ?tune.spls- new function
impute.nipals to impute missing values using the nipals
algorithm
- new function
tune.spca to tune the number of selected variables for
pca components
circosPlot now has methods for block.spls objects. It can now
handle similar feature names across blocks. It is also much more
customisable. See advanced arguments in ?circosPlot- new
biplot function for pca and pls objects. See
?mixOmics::biplot
plotDiablo now takes col.per.group (see #119)
April 2020
- weighted consensus plots for DIABLO objects now consider per-component
weights
March 2020
plotIndiv now supports (weighted) consensus plots for block
analyses. See the example in this
issueplotIndiv(..., ind.names=FALSE) warning
issue now fixed
January 2020
perf.block.splsda now supports calculation of combined AUCblock.splsda bug which could drop some classes with
near.zero.variance=TRUE now fixed
mixOmicsTeam/mixOmics documentation built on Nov. 4, 2024, 8:56 a.m.

This repository contains the R package which is hosted on
Bioconductor
and our stable and development GitHub versions.
Installation
(macOS users only: Ensure you have installed XQuartz first.)
From Bioconductor
The best way to install mixOmics is using Bioconductor. You can see
the landing page for the release version of mixOmics on Bioconductor
here.
Make sure you have the latest R version and the latest BiocManager
package installed following these
instructions.
## install BiocManager if not installed
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
## install mixOmics
BiocManager::install('mixOmics')
## load mixOmics
library(mixOmics)
From Github
Bioconductor versions are updated twice a year, between these updates
you can downlod the latest stable version of mixOmics from Github
using:
BiocManager::install('mixOmicsTeam/mixOmics')
You can also install the development version for new features yet to be widely tested:
BiocManager::install("mixOmicsTeam/mixOmics@development")
From Docker container
You can install our latest stable Github version of mixOmics via our
Docker container. You can do this by downloading and using the Docker
desktop application or via the command line as described below.
Note: this requires root privileges
1) Install Docker following instructions at https://docs.docker.com/docker-for-mac/install/
if your OS is not compatible with the latest version download an older version of Docker from the following link:
- MacOS: https://docs.docker.com/docker-for-mac/release-notes/
- Windows: https://docs.docker.com/docker-for-windows/release-notes/
Then open your system’s command line interface (e.g. Terminal for MacOS and Command Promot for Windows) for the following steps.
MacOS users only: you will need to launch Docker Desktop to activate your root privileges before running any docker commands from the command line.
2) Pull mixOmics container
docker pull mixomicsteam/mixomics
3) Ensure it is installed
The following command lists the running images:
docker images
This lists the installed images. The output should be something similar to the following:
$ docker images
> REPOSITORY TAG IMAGE ID CREATED SIZE
> mixomicsteam/mixomics latest e755393ac247 2 weeks ago 4.38GB
4) Activate the container
Running the following command activates the container. You must change
your_password to a custom password of your own. You can also customise
ports (8787:8787) if desired/necessary. see
https://docs.docker.com/config/containers/container-networking/ for
details.
docker run -e PASSWORD=your_password --rm -p 8787:8787 mixomicsteam/mixomics
5) Run
In your web browser, go to http://localhost:8787/ (change port if
necessary) and login with the following credentials:
username: rstudio password: (your_password set in step 4)
6) Inspect/stop
The following command lists the running containers:
sudo docker ps
The output should be something similar to the following:
$ sudo docker ps
> CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
> f14b0bc28326 mixomicsteam/mixomics "/init" 7 minutes ago Up 7 minutes 0.0.0.0:8787->8787/tcp compassionate_mestorf
The listed image ID can then be used to stop the container (here
f14b0bc28326)
docker stop f14b0bc28326
Contribution
We welcome community contributions concordant with our code of
conduct.
We strongly recommend adhering to Bioconductor’s coding
guide for
software consistency if you wish to contribute to mixOmics R codes.
Bug reports and pull requests
To report a bug (or offer a solution for a bug!) visit: https://github.com/mixOmicsTeam/mixOmics/issues. We fully welcome and appreciate well-formatted and detailed pull requests. Preferably with tests on our datasets.
Set up development environment- Install the latest version of R
- Install RStudio
- Clone this repo, checkout master branch, pull origin and then run:
install.packages("renv", Ncpus=4)
install.packages("devtools", Ncpus=4)
# restore the renv environment
renv::restore()
# or to initialise renv
# renv::init(bioconductor = TRUE)
# update the renv environment if needed
# renv::snapshot()
# test installation
devtools::install()
devtools::test()
# complete package check (takes a while)
devtools::check()
Discussion forum
We wish to make our discussions transparent so please direct your analysis questions to our discussion forum https://mixomics-users.discourse.group. This forum is aimed to host discussions on choices of multivariate analyses, as well as comments and suggestions to improve the package. We hope to create an active community of users, data analysts, developers and R programmers alike! Thank you!
About the mixOmics team
mixOmics is collaborative project between Australia (Melbourne),
France (Toulouse), and Canada (Vancouver). The core team includes
Kim-Anh Lê Cao - https://lecao-lab.science.unimelb.edu.au (University
of Melbourne), Florian Rohart - http://florian.rohart.free.fr
(Toulouse) and Sébastien Déjean -
https://perso.math.univ-toulouse.fr/dejean/. We also have key
contributors, past (Benoît Gautier, François Bartolo) and present (Al
Abadi, University of Melbourne) and several collaborators including
Amrit Singh (University of British Columbia), Olivier Chapleur (IRSTEA,
Paris), Antoine Bodein (Universite de Laval) - it could be you too, if
you wish to be involved!.
The project started at the Institut de Mathématiques de Toulouse in
France, and has been fully implemented in Australia, at the University
of Queensland, Brisbane (2009 – 2016) and at the University of
Melbourne, Australia (from 2017). We focus on the development of
computational and statistical methods for biological data integration
and their implementation in mixOmics.
Why this toolkit?
mixOmics offers a wide range of novel multivariate methods for the
exploration and integration of biological datasets with a particular
focus on variable selection. Single ’omics analysis does not provide
enough information to give a deep understanding of a biological system,
but we can obtain a more holistic view of a system by combining multiple
’omics analyses. Our mixOmics R package proposes a whole range of
multivariate methods that we developed and validated on many biological
studies to gain more insight into ’omics biological studies.
Want to know more?
www.mixOmics.org (tutorials and resources)
Our latest bookdown vignette: https://mixomicsteam.github.io/mixOmics-Vignette/
Different types of methods
We have developed 17 novel multivariate methods (the package includes 19 methods in total). The names are full of acronyms, but are represented in this diagram. PLS stands for Projection to Latent Structures (also called Partial Least Squares, but not our preferred nomenclature), CCA for Canonical Correlation Analysis.
That’s it! Ready! Set! Go!
Thank you for using mixOmics!

What’s New
March 2022
- bug fix implemented for Issue
#196.
perf()can now handle features with a(s)plswhich have near zero variance. - bug fix implemented for Issue
#192.
predict()can now handle when the testing and training data have their columns in different orders. - bug fix implemented for Issue
#178. If the
indYparameter is used inblock.spls(),circosPlot()can now properly identify the $Y$ dataframe. - bug fix implemented for Issue
#172.
perf()now returns values for thechoice.ncompcomponent whennrepeat$< 3$ whereas before it would just returnNAs. - bug fix implemented for Issue
#171.
cim()now can takepcaobjects as input. - bug fix implemented for Issue
#161.
tune.spca()can now handleNAvalues appropriately. - bug fix implemented for Issue
#150. Provided
users with a specific error message for when
plotArrow()is run on a(mint).(s)plsdaobject. - bug fix implemented for Issue
#122. Provided
users with a specific error message for when a
splsdaobject that has only one sample associated with a given class is passed toperf(). - bug fix implemented for Issue
#120.
plotLoadings()now returns the loading values for features from all dataframes rather than just the last one when operating on a(mint).(block).(s)plsdaobject. - bug fix implemented for Issue
#43. Homogenised
the way in which
tune.mint.splsda()andperf.mint.splsda()calculate balanced error rate (BER) as there was disparity between them. Also made the global BER a weighted average of BERs across each study. - enhancement implemented for Issue
#30/#34. The
parameter
verbose.callwas added to most of the methods. This parameter allows users to access the specific values input into the call of a function from its output. - bug fix implemented for Issue
#24.
background.predict()can now operate onmint.splsdaobjects and can be used as part ofplotIndiv().
July 2021
- new function
plotMarkersto visualise the selected features in block analyses (see https://github.com/mixOmicsTeam/mixOmics/issues/134) tune.splsnow able to tune the selected variables on bothXandY. See?tune.spls- new function
impute.nipalsto impute missing values using the nipals algorithm - new function
tune.spcato tune the number of selected variables for pca components circosPlotnow has methods forblock.splsobjects. It can now handle similar feature names across blocks. It is also much more customisable. See advanced arguments in?circosPlot- new
biplotfunction forpcaandplsobjects. See?mixOmics::biplot plotDiablonow takescol.per.group(see #119)
April 2020
- weighted consensus plots for DIABLO objects now consider per-component weights
March 2020
plotIndivnow supports (weighted) consensus plots for block analyses. See the example in this issueplotIndiv(..., ind.names=FALSE)warning issue now fixed
January 2020
perf.block.splsdanow supports calculation of combined AUCblock.splsdabug which could drop some classes withnear.zero.variance=TRUEnow fixed
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.


 -lightgrey.svg)
-lightgrey.svg)