README.md
In uc-bd2k/KEGGlincs: Visualize all edges within a KEGG pathway and overlay LINCS data [option]
title: "Introduction to KEGGlincs"
author: "Shana White"
date: "03/23/2017"
by Shana White
Installation:
#Make sure that the following bioconductor packages are installed
source("http://bioconductor.org/biocLite.R")
biocLite(c("hgu133a.db", "KEGGgraph", "KEGGREST", "KOdata"))
#Download package
biocLite("KEGGlincs")
#Load/activate package for use
library(KEGGlincs)
Overview of package

KEGGlincs is an R package that provides a seamless interface for viewing detailed versions of KEGG pathways in Cytoscape such that the exact relationships (edges) that exist between pathway elements (nodes) are visualized in a meaningful manner.
This package is intended to be used as a tool to accurately regenerate KEGG pathways with expanded edges that are marked-up (in terms of color or color+width) to portray specific edge attributes. Specifically,
- the edges are expanded to represent all relationships between genes encoded by the KGML file but not necessarily represented in the original [KEGG] pathway. This 'expansion' occurs either because:
- one node may represent many [often paralogous] genes.
- a node is of type 'group' and represents two or more genes that are part of the same functional complex.
- the edge attributes represented can be specified by the user as either:
- Functional - the exact type and [in some cases] subtype of the relationship defined by the KGML file.
- Data driven - based on quantitative experimental evidence for gene-gene relationships generated by experimental data. As an added feature and motivating example, this package gives users an opportunity to use LINCS L1000 knock-out data in order to analyze/visualize the pathway edges as they pertain to specific cell lines.
Aside from edge-specific annotation/mark-up, a unique feature of this package lies in the ability for users to generate graph objects in R and send them to Cytoscape for an interactive visualization experience via utilities developed by the cyREST team.
The following example showcases the ideas described above.
Example: p53 pathway
Original KEGG pathway:
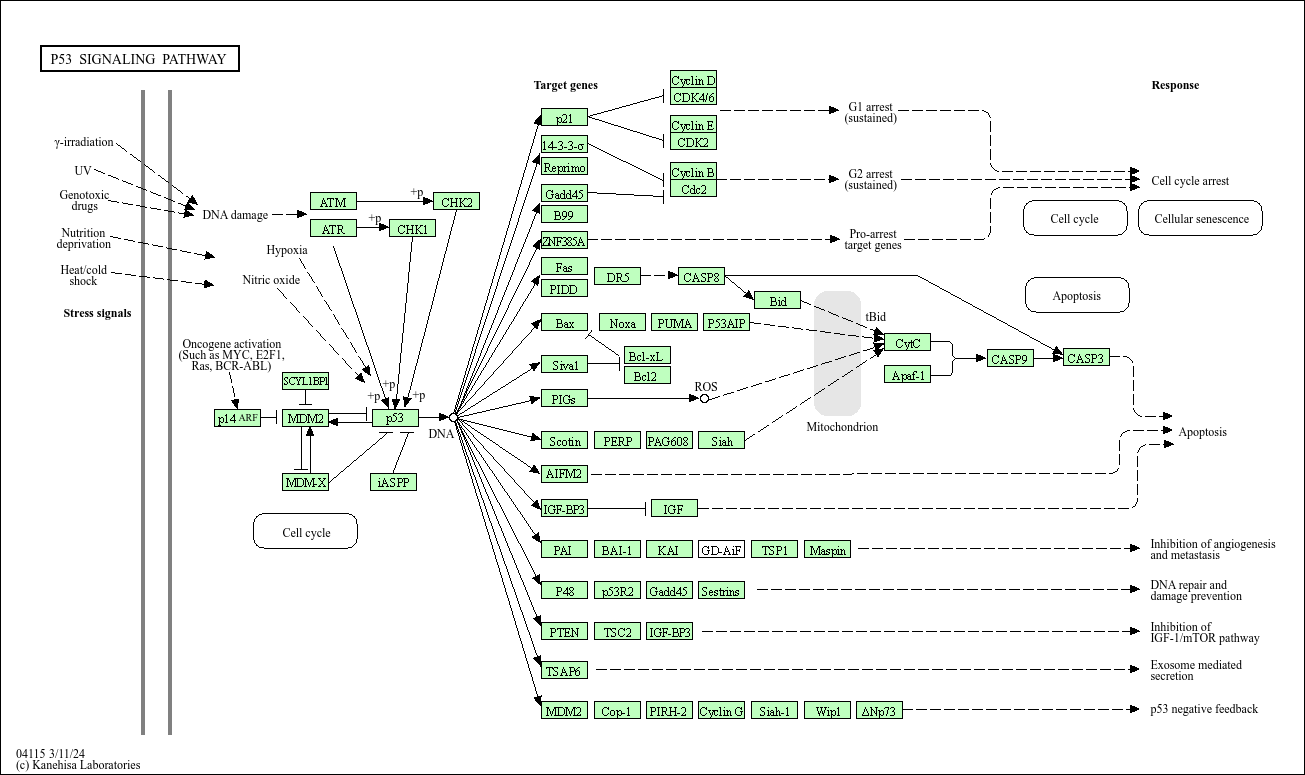
Using KEGGlincs without adding data:
Detailed pathway visualized in Cytoscape
- Explicitly shows all documented edges
- Color-coded to easily interpret edge type (i.e. activation, inhibition, etc.)
- Edge color key:

Note: An open Cytoscape session is required for pathway visualization; i.e. make sure that Cytoscape is open before running the following commands
KEGG_lincs("hsa04115")

Overlay pathway edges with LINCS data:
Pathway with edge attributes generated from LINCS L1000 knock-out data, refined by cell-line specific expression (default option, see below), and visualized in Cytoscape
- Explicitly shows all documented edges; edges that do not exist in the data appear but are not marked-up for interpretation [of significance].
- If baseline expression data is available for the selected cell-line, the pathway will be refined to only include edges between genes that are expressed in a given cell line. The nodes for unexpressed genes are colored gray.
- Widths: Magnitude of concordance for the edge in the selected cell-type
- Colors: Direction and significance for the edge in the selected cell-type (based on a modified odds-ratio score).
- Edge color key (+/- refers to direction of score; scores are considered significant if adjusted p-value is >= 0.05):

Pathway specific to PC3 cell-line (Prostate cancer)
KEGG_lincs("hsa04115", "PC3")

Pathway specific to HA1E cell-line (Immortalized normal kidney epithelial)
KEGG_lincs("hsa04115", "HA1E")

uc-bd2k/KEGGlincs documentation built on May 3, 2019, 2:13 p.m.
title: "Introduction to KEGGlincs" author: "Shana White" date: "03/23/2017"
by Shana White
Installation:
#Make sure that the following bioconductor packages are installed
source("http://bioconductor.org/biocLite.R")
biocLite(c("hgu133a.db", "KEGGgraph", "KEGGREST", "KOdata"))
#Download package
biocLite("KEGGlincs")
#Load/activate package for use
library(KEGGlincs)
Overview of package

KEGGlincs is an R package that provides a seamless interface for viewing detailed versions of KEGG pathways in Cytoscape such that the exact relationships (edges) that exist between pathway elements (nodes) are visualized in a meaningful manner.
This package is intended to be used as a tool to accurately regenerate KEGG pathways with expanded edges that are marked-up (in terms of color or color+width) to portray specific edge attributes. Specifically, - the edges are expanded to represent all relationships between genes encoded by the KGML file but not necessarily represented in the original [KEGG] pathway. This 'expansion' occurs either because: - one node may represent many [often paralogous] genes. - a node is of type 'group' and represents two or more genes that are part of the same functional complex. - the edge attributes represented can be specified by the user as either: - Functional - the exact type and [in some cases] subtype of the relationship defined by the KGML file. - Data driven - based on quantitative experimental evidence for gene-gene relationships generated by experimental data. As an added feature and motivating example, this package gives users an opportunity to use LINCS L1000 knock-out data in order to analyze/visualize the pathway edges as they pertain to specific cell lines.
Aside from edge-specific annotation/mark-up, a unique feature of this package lies in the ability for users to generate graph objects in R and send them to Cytoscape for an interactive visualization experience via utilities developed by the cyREST team.
The following example showcases the ideas described above.
Example: p53 pathway
Original KEGG pathway:

Using KEGGlincs without adding data:
Detailed pathway visualized in Cytoscape
- Explicitly shows all documented edges
- Color-coded to easily interpret edge type (i.e. activation, inhibition, etc.)
- Edge color key:
Note: An open Cytoscape session is required for pathway visualization; i.e. make sure that Cytoscape is open before running the following commands
KEGG_lincs("hsa04115")

Overlay pathway edges with LINCS data:
Pathway with edge attributes generated from LINCS L1000 knock-out data, refined by cell-line specific expression (default option, see below), and visualized in Cytoscape
- Explicitly shows all documented edges; edges that do not exist in the data appear but are not marked-up for interpretation [of significance].
- If baseline expression data is available for the selected cell-line, the pathway will be refined to only include edges between genes that are expressed in a given cell line. The nodes for unexpressed genes are colored gray.
- Widths: Magnitude of concordance for the edge in the selected cell-type
- Colors: Direction and significance for the edge in the selected cell-type (based on a modified odds-ratio score).
- Edge color key (+/- refers to direction of score; scores are considered significant if adjusted p-value is >= 0.05):
Pathway specific to PC3 cell-line (Prostate cancer)
KEGG_lincs("hsa04115", "PC3")

Pathway specific to HA1E cell-line (Immortalized normal kidney epithelial)
KEGG_lincs("hsa04115", "HA1E")

Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
