In ramiromagno/gwasrapidd: 'REST' 'API' Client for the 'NHGRI'-'EBI' 'GWAS' Catalog
knitr::opts_chunk$set(
collapse = TRUE,
# comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
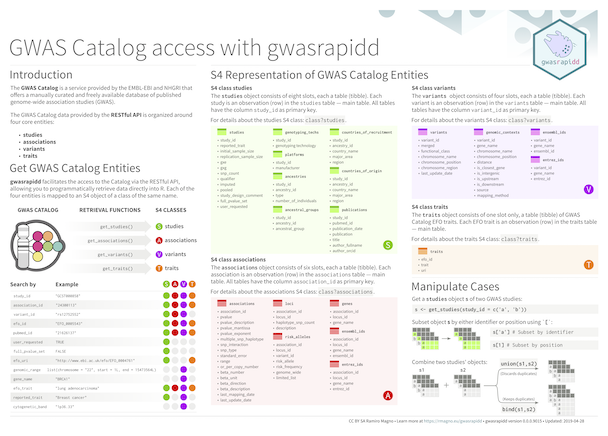
gwasrapidd 
The goal of {gwasrapidd} is to provide programmatic access to the NHGRI-EBI
Catalog of published genome-wide association
studies.
Get started by reading the
documentation.
Installation
Install {gwasrapidd} from CRAN:
install.packages("gwasrapidd")
Cheatsheet
Example
Get studies related to triple-negative breast cancer:
library(gwasrapidd)
studies <- get_studies(efo_trait = 'triple-negative breast cancer')
studies@studies[1:4]
Find associated variants with study r studies@studies$study_id[1]:
variants <- get_variants(study_id = 'GCST002305')
variants@variants[c('variant_id', 'functional_class')]
Citing this work
{gwasrapidd} was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
citation('gwasrapidd')
Code of Conduct
Please note that the {gwasrapidd} project is released with a Contributor Code
of Conduct. By contributing
to this project, you agree to abide by its terms.
Similar projects
- Bioconductor R package gwascat by Vincent J Carey:
https://www.bioconductor.org/packages/release/bioc/html/gwascat.html
- Web application PhenoScanner V2 by Mihir A. Kamat, James R. Staley, and
others:
http://www.phenoscanner.medschl.cam.ac.uk/
- Web application GWEHS: Genome-Wide Effect sizes and Heritability Screener by
Eugenio López-Cortegano and Armando Caballero:
http://gwehs.uvigo.es/
Acknowledgements
This work would have not been possible without the precious help from the GWAS
Catalog team, particularly Daniel
Suveges.
ramiromagno/gwasrapidd documentation built on Jan. 3, 2024, 10:21 p.m.
knitr::opts_chunk$set( collapse = TRUE, # comment = "#>", fig.path = "man/figures/README-", out.width = "100%" )
gwasrapidd 
The goal of {gwasrapidd} is to provide programmatic access to the NHGRI-EBI
Catalog of published genome-wide association
studies.
Get started by reading the documentation.
Installation
Install {gwasrapidd} from CRAN:
install.packages("gwasrapidd")
Cheatsheet
Example
Get studies related to triple-negative breast cancer:
library(gwasrapidd) studies <- get_studies(efo_trait = 'triple-negative breast cancer') studies@studies[1:4]
Find associated variants with study r studies@studies$study_id[1]:
variants <- get_variants(study_id = 'GCST002305') variants@variants[c('variant_id', 'functional_class')]
Citing this work
{gwasrapidd} was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
citation('gwasrapidd')
Code of Conduct
Please note that the {gwasrapidd} project is released with a Contributor Code
of Conduct. By contributing
to this project, you agree to abide by its terms.
Similar projects
- Bioconductor R package gwascat by Vincent J Carey: https://www.bioconductor.org/packages/release/bioc/html/gwascat.html
- Web application PhenoScanner V2 by Mihir A. Kamat, James R. Staley, and others: http://www.phenoscanner.medschl.cam.ac.uk/
- Web application GWEHS: Genome-Wide Effect sizes and Heritability Screener by Eugenio López-Cortegano and Armando Caballero: http://gwehs.uvigo.es/
Acknowledgements
This work would have not been possible without the precious help from the GWAS Catalog team, particularly Daniel Suveges.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.